Generate ideal helices and strands
This script shows how to generate idealized helices and strands from a sequence.
Introduction
For certain purposes, such as writing tests for cctbx, it is useful to generate a model of idealized secondary structure, such as helices and beta strands.
Code
The script needs the following imports:
from __future__ import absolute_import, division, print_function
from mmtbx.secondary_structure.build import ss_idealization as ssb
The first line makes sure that the script runs with both Python2 and Python3.
The second import from the mmtbx/secondary_structure module makes available the function used to generate ideal secondary structure.
Then we create an ideal helix from a sequence, here we use the example sequence "ILMKARNDWYV":
ph_helix = ssb.secondary_structure_from_sequence(ssb.alpha_helix_str,"ILMKARNDWYV")
The secondary_structure_from_sequence function creates a new pdb_hierarchy object. The first argument is a string in PDB format (pdb_str) that has the geometry of the secondary structure. The following strings are available in the code; in parenthesis is the name of the variable in the code:
- Alpha helix (
alpha_helix_str) - 3-10 helix (
a310_helix_str) - Pi-helix (
pi_helix_str) - Beta strand (
beta_pdb_str)
The second argument is the sequence, in form of a string of one-letter codes. It can be an explicit sequence, such as "ILMKARNDWYV", or in form of "A"*20 for 20 alanine residues.
Once we have the new pdb_hierarchy for the idealized helix, we can save it as PDB file:
ph_helix.write_pdb_file(file_name="m-helix.pdb")
Similarly, we can create an ideal beta strand:
ph_strand = ssb.secondary_structure_from_sequence(ssb.beta_pdb_str,"ILMKARNDWYV")
ph_strand.write_pdb_file(file_name="m-strand.pdb")
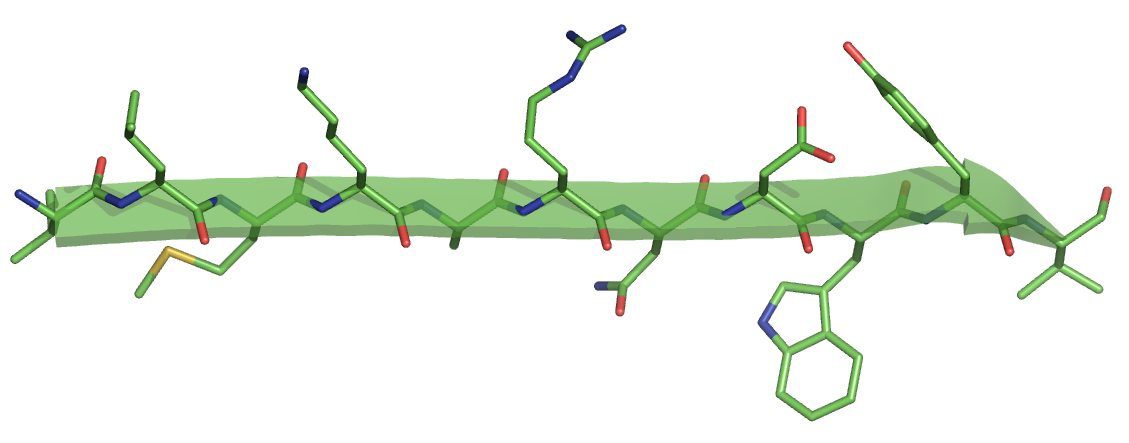
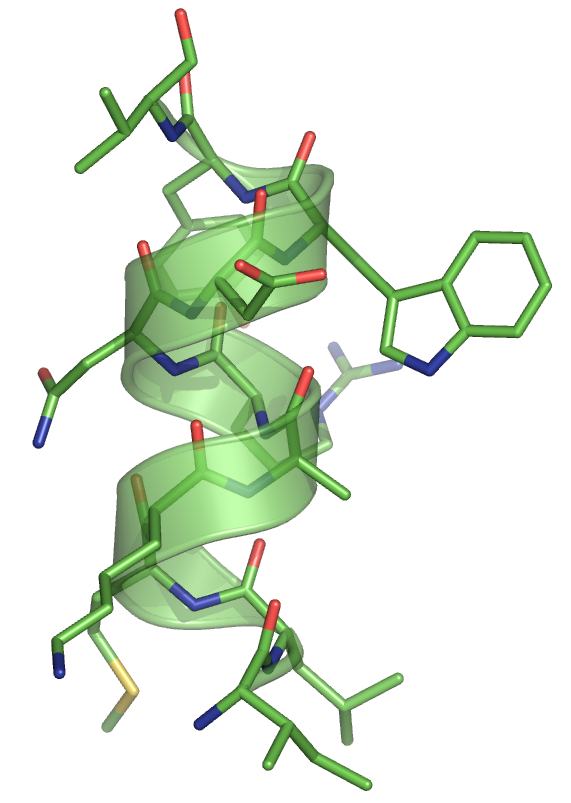
Example
For the sequence used in our example ("ILMKARNDWYV"), we get the ideal alpha helix below.
For the same sequence, we get the follwing strand. Note the peptide N-H and C=O groups pointing to the top and the bottom.