Replacing a segment of a model
Replace a part of a model by swapping in a segment from a similar model and learn how to combine models to create a composite model.
Setting up example data
First, let's set up the example data (described in more detail here):
Get the files from the Phenix regression directory and change into the new folder:
phenix.setup_tutorial tutorial_name=model-building-scripting
cd model-building-scriptingType phenix.python to start up Python with the Phenix environment all set up:
phenix.pythonSet up the high level objects, so we can start our task:
from iotbx.data_manager import DataManager # Load in the DataManager
dm = DataManager() # Initialize the DataManager and call it dm
dm.set_overwrite(True) # Overwrite files with the same name
model = dm.get_model("structure_search_target.pdb") # read in a model
mmm = dm.get_map_model_manager( # getting a map_model_manager
model_file = "structure_search_target.pdb",)
Replacing a segment
Let’s replace a part of a model by finding a similar model in the PDB and swapping in the segment from that model. This procedure is very similar to fitting a loop, but we are going to use a segment right from the PDB to fill in our loop instead of trying to build it from scratch.
The map_model_manager mmm knows how to generate a model-based map from a model. Let’s create a map at a resolution of 3 Å and specify that this is a cryo-EM map:
mmm.generate_map(d_min=3) # generate map to resolution of 3 A
mmm.set_experiment_type('cryo_em') # this is a cryo-EM map
Now we are ready to get a model_building object and to specify that we want to use the ‘structure_search’ method to replace a segment. Just to see how it works, we’ll turn on debugging (prints more output too) and set the thoroughness of the run to ‘quick’:
build = mmm.model_building() # get a model-building object
build.set_defaults( # set some defaults
thoroughness='quick', # quick run
debug=True,) # turn on debugging output
We can save and write out the starting model before we modify it:
model_before_replace = build.model().deep_copy() # save model
dm.write_model_file(build.model(), 'model_before_replace.pdb')# starting model
We will replace residues 22-34 (show in orange in the figure below).
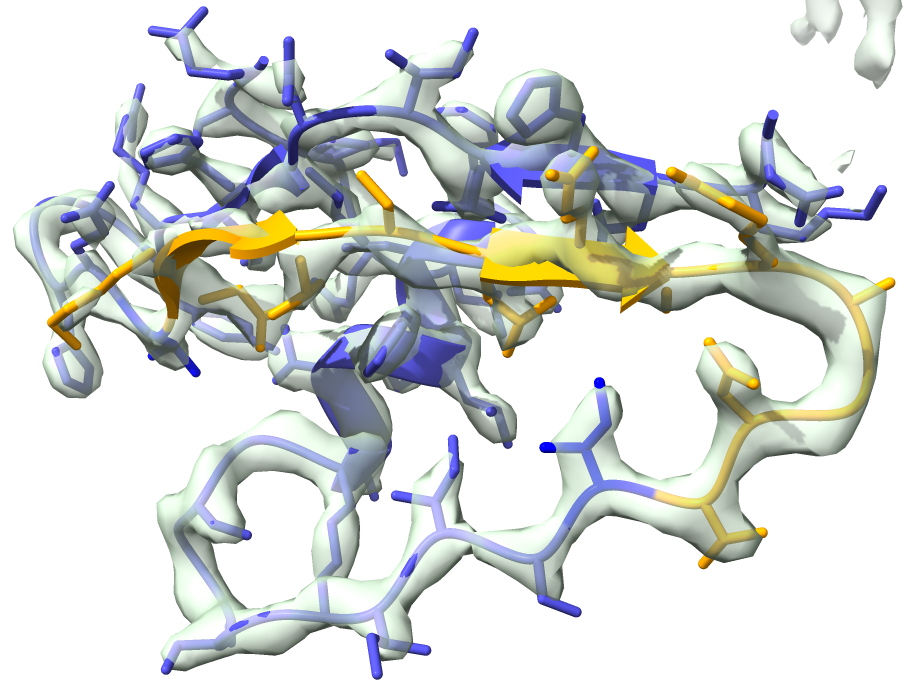
We can now run the segment replacement:
replace_segment_model=build.replace_segment( # replace a segment
chain_id="A", # chain to work on
last_resno_before_replace=21, # just before replace
first_resno_after_replace=35, # just after replace
refine_cycles=1,) # cycles of refinement
And we can insert the new segment, get a new map_model_manager, and write out the results:
build.insert_fitted_loop() # inserts loops or segments
mam=build.as_map_model_manager() # get a map_model_manager to write out with
mam.write_model('model_with_replacement.pdb') # the model
mam.write_map('map_for_replacement.ccp4') # the map
Have a look at the starting model (model_before_replace.pdb), the model with a replacement segment (model_with_replacement.pdb), and the map (map_for_replacement.ccp4).
Combining Models
Let’s combine several models and create a new model that has the best parts of each. We can use two models that we just worked with (build.model() and model_before_replace) along with the map we just used. The working model_building object contains model_with_replacement.pdb and the working map.
Just to see how it works, let’s create two overlapping models: residues 6-25 from the starting model, model_before_replace, and residues 20 - 60 from the model with a segment replaced (in build.model()):
model_1 = model_before_replace.apply_selection_string( # apply a selection
'resseq 6:25' ) # keep only residues 6 through 25
model_2 = build.model().apply_selection_string( # apply a selection
'resseq 20:60' ) # keep only residues 20 through 60
The overlapping selections for the two models are shown below; model_1 is blue, model_2 is red, the overlappting segment is encircled.
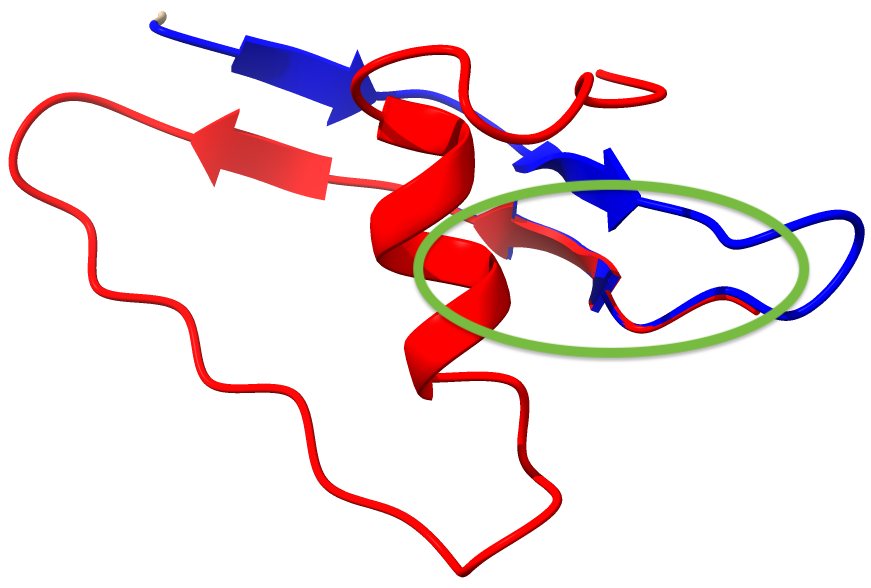
We can make sure this worked:
model_1.get_hierarchy().overall_counts().n_residues # residues in model_1
model_2.get_hierarchy().overall_counts().n_residues # residues in model_2
Now we can combine the overlapping models with:
new_model = build.combine_models( # combine models
model_list = [model_1, model_2]) # models to combine
We can write out the new combined model with:
dm.write_model_file(new_model, 'combined_model.pdb') # write out new model
Have a look at model_before_replace.pdb, model_with_replacement.pdb, and combined_model.pdb. The combined model has residue 20 from model_1 and residue 30 from model_2.