Assign a sequence to a model
Learn how to to assign a sequence to a model or to guess the sequence from the density.
Setting up example data
First, let's set up the example data (described in more detail here):
Get the files from the Phenix regression directory and change into the new folder:
phenix.setup_tutorial tutorial_name=model-building-scripting
cd model-building-scriptingType phenix.python to start up Python with the Phenix environment all set up:
phenix.pythonSet up the high level objects, so we can start our task:
from iotbx.data_manager import DataManager # Load in the DataManager
dm = DataManager() # Initialize the DataManager and call it dm
dm.set_overwrite(True) # Overwrite files with the same name
Assigning a sequence
The model_building object has a tool that examines the density in a map at the positions of each side-chain in a model and tries to figure out what side chain belongs there. This can be used to assign a sequence to a model or to guess the sequence from the density. Let’s use it to assign a sequence to a model.
Use the DataManager to read in a model and map from our working directory:
mmm = dm.get_map_model_manager( # getting a map_model_manager
model_file="short_model_main_chain_box.pdb", # model file
map_files="short_model_box.ccp4") # map file
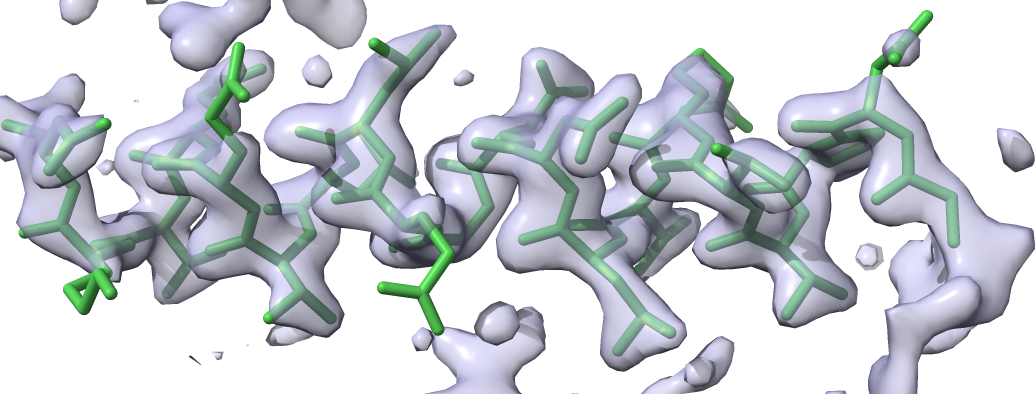
The model consists of alanine residues:
We need one extra file this time, a sequence file. Let’s read our sequence as an object from seq.dat:
seq_object=dm.get_sequence("seq.dat") # read sequence object from seq file
Our sequence as text is available from the sequence object:
sequence_as_text = seq_object[0].sequence # get simple text sequence
We can set the resolution and experiment type and get a model_building object:
mmm.set_resolution(3.0) # working (nominal) resolution of map
mmm.set_experiment_type('xray') # define experiment type
build = mmm.model_building() # get a model-building object
Now we are all set. We can just say:
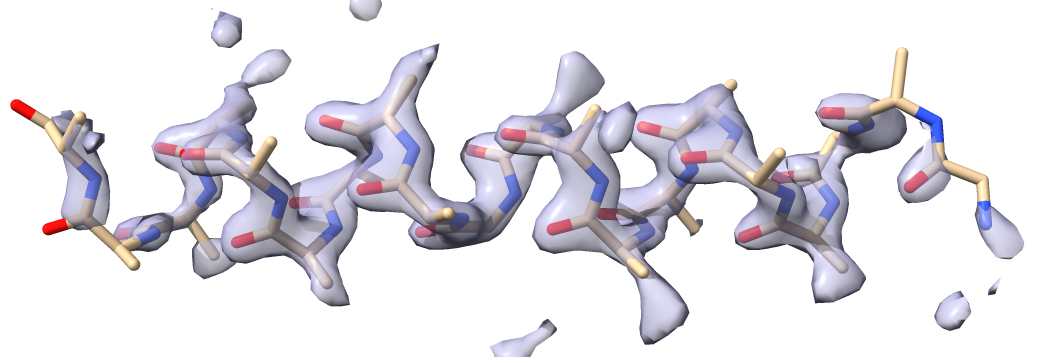
build.sequence_from_map(sequence=sequence_as_text) # get sequence from the map
...and our working model will be replaced by a model with sequence assigned (if we are lucky). We can write out the new model and compare it with the original in Coot or Chimera along with the map in short_model_box.ccp4:
dm.write_model_file(build.model(), 'sequence_assigned.pdb') # write out model