HKLviewer
Learn how to visualize the reflections of your diffraction dataset.
Author: Robert D. Oeffner
Introduction
The HKLviewer lets you inspect diffraction data sets to understand possible issues that may hamper structure solution.
The HKlviewer is available from a CCTBX installation. In the command line, type cctbx.HKLviewer in a system shell. Alternatively double click the file icon of CCTBX\build\bin\cctbx.HKLviewer in a file manager. From the command line, add the name of a reflection file to open it immediately.
If you find HKLviewer useful in your work please cite Computational Crystallography Newsletter (2021). 12, 15-25 in your publication.
Bug reports or queries
Please submit bug reports or questions to rdo20@cam.ac.uk or cctbx@cci.lbl.gov. For suggested enhancements to the HKLviewer feel free to make a pull request on https://github.com/cctbx/cctbx_project.
Opening a file
A reflection file can be openend from the "File | Open" menu item. Alternatively the filename can be specified as an argument on the command line where the CCTBX environment has been invoked:
cctbx.HKLviewer 3EIL.mtzThe types of reflection files that can be loaded are .mtz, .hkl, .cif and .sca files (assuming the space group dimension is present in the file).
Display reflection data
After a file has been loaded, a particular data set can be displayed by double-clicking the row with the corresponding label in the upper left table. The viewer will display reflections in 3 dimensions according to their reciprocal space coordinates (h,k,l values) and with respect to the space group dimension of the crystal. The size and the colour of the sphere of each reflection depends on its data value.
Display uncertainties (sigma)
If a dataset contains uncertainties (sigma) in addition to diffraction data, say it is labelled as "F,SIGF", then they can be displayed with a right click on the respective row in the upper-left table. A menu will appear where the second item will be "Display sigmas of F,SIGF". Selecting that item will display the data in the viewer with the colour mapping and the size of the reflection spheres now being mapped according to sigma values in the dataset.
Colour mapping
The reflections are coloured according to colour gradient maps of the Python module matplotlib. The colour mapping can be changed with a mouse-click on the upper left colour chart in the viewer area of a displayed dataset. This will bring up the Colour gradient maps dialog as show below:

A colour gradient map is an array of rgb values that usually varies smoothly between adjacent array elements. The mapping between a data value, x, and a colour gradient map is done with a power law:
\begin{equation} f(x) = ((x-x_{min})/(x_{max} - x_{min}))^p \end{equation} where \(x_{min} \) and \(x_{max}\) are the minimum and maximum data values found in the data set. The function \(f(x)\) as defined will take on values between 0 and 1. It is then mapped onto the array values of the colour gradient map chosen by the user. With \(x\) varying linearly between \(x_{min} \) and \(x_{max}\) and for \(p < 1\) the first part of the colour gradient array will be emphasized. If \(p > 1\) it will be the last part of the colour gradient array. Some data types such as intensities are better viewed with a value of \(p\) that is smaller than 1. Other data types, like figure of merits are best viewed with a linear mapping between colours and the data values, i.e. p should be set to 1. Below is an illustration of how different power factors affects the resultant colour mapping for brg being the chosen colour gradient map.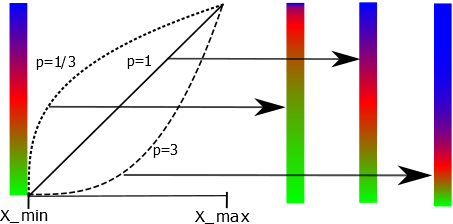
Displaying map coefficients
For data with map coeffficients or phases only the phase value (modulo 360) is used for the colour mapping. In this case it makes sense to choose a circular colour mapping, i.e. one where the colour at one end of the array equals the colour at the beginning of the array. Amongst the maps available from matplotlib these are hsv and gist_rainbow. For data with map coefficients or phases the power law mapping is fixed at p=1.
Clicking on a desired colour gradient in the Colour gradient maps dialog will instruct the HKLviewer to use this colour mapping for the displayed data or sigma values. This choice is used for all datasets of the same type as the currently shown one. This means that the colour mapping chosen for, say a dataset of amplitudes will cause any other dataset of amplitudes to use the same colour mapping.
Expansion
Processed reflection data usually are stored with only the reflections that are unique under symmetry operations, meaning that two different reflections are not mapped to the same reflections after symmetry operations have been applied. This subset of the recorded reflections are for convenience stored in the same wedge of the sphere of all reflections. For the P1 space group the wedge will be shaped as a half-sphere given that reflections present in such a wedge are the minimum number of reflections necessary for a complete (up to a given resolution) P1 data set, disregarding Friedel mates. For a high symmetry spacegroup such as a cubic spacegroup the wedge of reflections will be much smaller and shaped like a parsnip with sharp edges.
There are situations where it is preferable to inspect reflection data as if it had been stored as P1 data including Friedel mates. This is done in the HKLviewer by ticking the "Expand reflections" checkbox on the "Expansion" tab. This will apply the spacegroup rotations to each reflectionand display it accordingly. If the reflection data file does contain reflections that are not unique under symmetry only the option of P1 expansion will be available.
It is also possible to display "Missing reflections". These are the reflections that would have been present in a given wedge of reflections up to the resolution limit of the data set but have been lost during measurement, say due to radiation damage of the crystal or a shading beamstop.
Below is an example of showing only the missing reflections for the data set 2B18 expanded to P1 including Friedel pairs:

Slicing reflections
It is often useful to look at a slice through the displayed reflections. It is most informative to first expand the reflections tot he full sphere on the "Expansion" tab. Reflections can be sliced with a clip plane parallel to the screen that allows for arbitrary orientation of the reflections by the user. This can be useful when examining reflections governed by a twin operator where the axis of rotation is not necessarily aligned with either one of the reciprocal unit cell axes.
Below are reflections of the data set 2B18 displayed with a clip plane normal to the HKL vector (2,0,-5) pointing out of the screen at a distance of 5.5 from the origin.

Reflections can also be shown as just a single plane of reflections restricted to the ones where one of the axes h,k,l has a fixed value as shown below.

Sizing the displayed reflections
The size of each displayed reflections is a function of its associated data value. Similar to colour mapping of data values a function is employed that produces a radius, \(r\), dependent on the data value, \(x\), and a power scale factor, \(p\).
\begin{equation} r(x) = ((x-x_{min})/(x_{max} - x_{min}))^p \end{equation} where \(x_{min} \) and \(x_{max}\) are the minimum and maximum data values found in the data set. The power scale factor can either be selected by the user or be determined atuomatically.
For data sets of amplitudes or intensities where there often are more orders of magnitude difference between the smallest and the largest data values it can sometimes be helpful to adjust the power scale factor to either make all reflections the same size, \(p=0\), to illustrate what was actually recorded. Alternatively, to get a better view of the strength of the data an automatic power scale factor can be selected which will render the weakest reflections ten times smaller than the strongest reflections. Selecting a power scale factor of 1 for amplitude or intensity data is rarely useful as typically the data set is dominated by a handful of very strong low resolution reflections. But it may serve for illustrating the presence of strong anisotropy or tNCS modulation in a data set.
Binning
A data set can be sorted into bins according to resolution and if more data sets are present in the reflection file it can also be sorted according to values in those data sets. The number of bins desired can be set from the spin control to a maximum of 40. The HKLviewer will attempt to divide the currently displayed data into bins having about the same number of reflections in each bin. This can be overwritten by entering a preferred threshold for a particular bin row under the "lower bin value" column.
In the figure below 6 bins of resolution have been created for the FP,SIGFP dataset.

Changing bin thresholds
If one were to change the bin threshold for 4th bin from 2.27 to 2.1Å this is done by entering the new value in its place.

Entering return on the keyboard processes the new value and computes new number of reflections in the bin and adjacent ones.

The reflections in this bin can be visualised by deselecting reflections in all other bins in the opacity column.

Data can alsso be binned against other data values by selecting the desired data value for binning from the drop-down list.

Below is the data set FP,SIGFP binned according to the sigmas found in the data set but only displayed for the data values with sigmas larger than 5.42. This is done by un-ticking all the checkboxes but the last one in the opacity column.

Data can also be made semi transparent by entering a value between 0 and 1 on the opacity column. This may be useful when showing vectors that are shorter than the radius of the sphere of reflections, i.e. the highest resolution shell. Beware that implementations of WebGL can often cause shading artifacts of objects when displayed semitransparently. If opacity is set below 0.3 reflections will not respond to hovering or clicking with the mouse.
Vectors
On the Vectors tab various vectors can be displayed in reciprocal space. These are the rotation axes given by the space group, any tNCS vector specified in the file header as well as vectors defined by the user either in reciprocal or in real space fractional coordinates. In the HKLviewer reciprocal space or real space vector are defined with (0,0,0) as their origin so only their end coordinates are listed in the table.

Above is an example of the viewer showing the reflections of a dataset together with the rotation axes as defined by the spacegroup. A tNCS vector has also been drawn. If only one vector is drawn the reflections can be oriented with respect to the vector being either parallel or perpendicular to the screen. It is then possible to also rotate the reflections around that vector which can be helpful for examining features in the dataset such as tNCS modulation or twin laws.
User defined vectors
In addition to rotation axes given by the space group it is possible to explicitly enter a rotation operator and show its rotation axes. This is done by overwriting the "new name" field in the "draw" column of the table of rotation operators or vectors with a name of choosing, say "some_rot".

For the dataset 2QCW with the spacegroup P31 2 1 the reflections are displayed below. Although not part of the spacegroup we can see that it is possible to apply a 6-fold rotation along the l-axis and have the reflections unambiguously transformed into new ones with the operation "-k,h+k,l". We enter that into the rotation column.

HKLviewer will parse the operator when the enter key is pressed. If parsing succeeds the row in the table will become read-only and the name specified in the draw column will have "n-fold_" prefixed to it, where n is the number of times to apply this operation to achieve the identity operator.

Rotation operators should be commensurate with the spacegroup. However, some leeway is allowed although grossly mismatching rotation operators will be refused with an error message. When displaying only one rotation vector the symmetry related reflections for a given reflection will be highlighted with vectors and tooltips when right-clicking on a particular reflection. This is shown below where a slice has been made at l=0 and reflections have been expanded to P1 and Friedel mates. This is useful when inspecting twin related reflections as discussed in the tutorial section.

It is also possible to enter a reciprocal space vector. To do this first overwrite the "new name" field in the "draw" column with a chosen name, say "myHKL". Then enter the reciprrocal space vector in the same row under the "as hkl" column.

When the HKLviewer parses the coordinates the row will become read-only and the coordinates encapsulated by brackets.

Real space fractional vectors may also be entered in the same manner as for reciprocal space vectors. Enter those in the "as abc" column.

The unit cells for real space and reciprocal space can also be drawn for the sake of locating the tNCS vector or rotation operators. in the unit cell. Below is the 2QCW data set displayed with its realspace unit cell, the 6-fold rotation axis, the myHKL and the myABC vectors defined above.

Making a new dataset
A new dataset can be created from one or two existing dataset that are present in the file loaded into the HKLviewer. The way to create a new dataset is from python scripting. The datasets present in the HKLviewer are stored as cctbx.miller.array python data types. The cctbx.miller.array module provides a rich API of functions for crystallography which makes many crystallographic computational tasks simple to do from python scripting.
To get started, right-click a dataset in the upperleft table in the HKLviewer which you want to use for creating a new dataset. This invokes a context menu. Then select the "Make a new dataset from this dataset and another dataset" menu item. This invokes the "Create a new reflection dataset" dialog.

In the textbox a python expression that defines the new dataset, newarray, can be entered. Below that, a label for the dataset should also be entered. Pressing the "Compute new reflection dataset" button then parses and executes the python expression. If succesful the new dataset with the specified column label will appear at the bottom of the table of existing datasets and automatically be displayed in the viewer. For convenience newarray is initialised to a copy of array1 (here representing "I,SIGI").
Variables
The following variables are available for a python expression submitted in the "Create a new reflection dataset" dialog:
| Variable or member function | Type | Description |
|---|---|---|
| newarray | cctbx.miller.array | The new dataset that is computed from the python script |
| array1 or array2 | cctbx.miller.array |
array1 is the variable name of dataset that was right-clicked in the upperleft table of dataset. array2 is the variable name of the dataset that optionally can be selected from the dropdown list |
| newarray._data, array1.data() or array2.data() | flex.double, flex,complex, flex.int or flex.hendrickson_lattman | Observed data values, figure of merits, map coefficients, Rfree or other column values present in the reflection file. |
| newarray._sigmas, array1.sigmas() or array2.sigmas() | flex.double or None if not provided | Observed sigma values associated with the data values. |
| array1.indices() or array2.indices() | flex.miller_index | HKL indices of each reflection. If array2 has been selected for the computation as well, the indices will be the common subset of indices of both datasets. |
| dres | flex.double | Resolution of each HKL index |
The dres variable is provided as a shorthand for array1.unit_cell().d(array1.indices()).
Examples of creating a new dataset
When creating a new dataset copies, the objective is to define newarray as a new cctbx.miller.array object or to assign its attributes newarray._data and newarray._sigmas to flex arrays computed in the script. The newarray is initalised to the dataset that was right-clicked for invoking the "Create a new reflection dataset" dialog. So the newarray.indices attribute needs not specifying. The array1 and array2, of the selected datasets are available for scripting in the "Create a new reflection dataset" dialog. The data(), sigmas() and indices() member functions of array1 and array2 can then be used for composing the new dataset. A few examples are given in the table below:
| Objective | Python script to be entered |
|---|---|
| Calculate normalized structure factors, E, from amplitude, F, or intensity, I, dataset. | newarray = array1.normalize() |
| Calculate I/SigI for an intensity dataset. |
dat = array1.data() / array1.sigmas() newarray._data = dat newarray._sigmas = None newarray.set_observation_type(None) |
| Multiply data values in one dataset with the squareroot of data values in another dataset |
dat = array1.data() * flex.sqrt( array2.data()) newarray._data = dat newarray._sigmas = None newarray.set_observation_type(None) |
| French-Wilson massage intensity data values. | newarray = array1.french_wilson() |
The important thing to bear in mind is that the newarray._data and optionally newarray._sigmas should be of type flex arrays and not scalar values, list or tuples. Many mathematical functions have been overloaded for flex arrays. So when computing the squareroot, the absolute value or raising all elements of a flex double array to a certain power use flex.sqrt(array1.data()) etc. Avoid using a for-loop or list comprehension over all elements in a flex array in the python script.
Further API details
There are many more functions available from the cctbx.miller.array class. Details about flex arrays can be found here and here.
Saving datasets
Once new datasets have been created it is possible to save these from the File menu by clicking the "Save Reflection File" item. Supported file formats are MTZ and CIF. If a reflection file contains unmerged data with multiple values for one or more indices each dataset will be saved in separate CIF files. Currently it is not possible to save unmerged data in the MTZ format.
Table of datasets
With a right click on the upper-left table of datasets in the reflection data file individual datasets can be tabulated from the context menu. First highlight the datasets of interest. Then click on "Show a table of

The table can be sorted according to the values of any available dataset, to the resolution. Double clicking a row in the table will cause the viewer to zoom in on that particular reflection with a red mesh

Similarly will a right-click on a displayed reflection in the viewer retrieve and highlight that reflections data value in the table.
The tabulated datasets will remain unchanged when the displayed reflections are transformed by expanding to Friedel mates, P1 or lower symmetry spacegroups. This also means that double clicking a symmetry copy generated from expanding the dataset to P1 or Friedel mates will only retrieve the original HKL index and its data value in the table.